Project Goal
To build the foundation required to connect a plant’s phenotype to its genotype and environmental conditions using computational methods, software, and data.
Project Summary
Tony Kusalik, Matthew Links, Michael Horsch and their team of researchers are using computational methods to map out the relationship between a plant’s phenotype (observable physical characteristics), its genotype (DNA structure), and the environment. This is a difficult task, as the plant genome is complex and involves many different contributing factors including in the microbial community surrounding its roots and foliage. To put the problem into perspective, consider that humans have diploid genetics with two sets of chromosomes. Plants are much more genetically complex with many crops being polyploid (for example, canola has six copies for every gene).
Researchers in this project have begun their work using simple cases and data. For example, they are using data from the model plant Arabidopsis as it has a smaller genome and is very similar to canola. Time and environment are also being de-emphasized in the initial phase of the project. Currently, the team is developing software artifacts that can be used later on. Later phases of the project will incorporate more complex information and data sets, as well as environmental information.
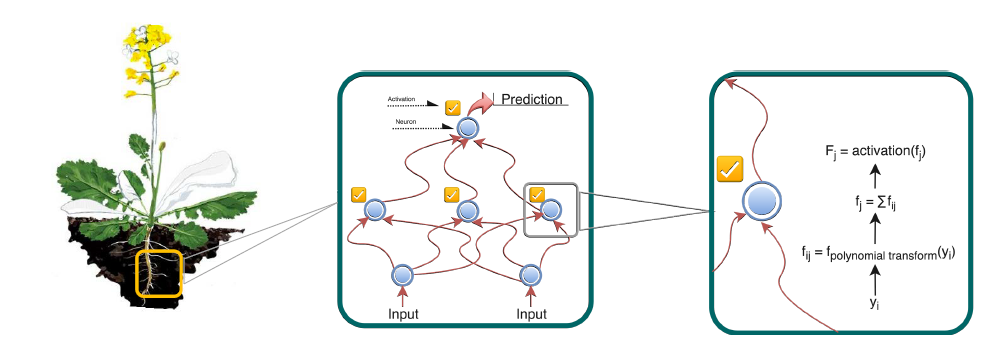
Project Results to Date
With so many potential genetic features contributing to a plant’s physical characteristics, in addition to environmental factors such as weather, soil, and microbes, it is challenging to determine which of these factors contribute significantly to plant features such as yield, seed color, or tolerance to drought and diseases.
To meet this challenge, the first step of this research is feature selection. This involves identifying the important features and removing features that are not as informative, using statistics and theory that deals with the limits and efficiency of information processing.
Following feature selection, researchers mine these features for associations between phenotype and genotype using Genome Wide Association Studies (GWAS), Deep Learning (DL), and Probabilistic Graphical Models (PGM).
- Genome Wide Association Studies are the standard approach to determining associations between genetics and traits, and form the baseline that other methods are compared to. GWAS is an existing technology often used with human genetics.
- Deep Learning involves various machine learning techniques that start with data, model it, and make predictions. It is used very successfully in applications such as speech recognition and face recognition. If there is a pattern in the data, then the Deep Learning procedure should be able to model it without any prior insight about that model.
- A Probabilistic Graphical Model is a machine learning method that involves modelling features using a combination of probability, statistics, and graph theory, with an aim to find the associations between variables and features which comprise the data. This is different from Deep Learning, because the model can be inspected by human biologists to assess if it makes sense.
Project 3.3 is also working to select a common language or descriptions – ontologies – that P2IRC researchers from diverse backgrounds can use to describe scientific ideas, results, and concepts related to their research.
Practical Applications
- One of the largest benefits of this research is that it will provide data analysis tools to inform plant breeders and their breeding programs. It will show where the breeding program will benefit the most, and which direction it should take. It may also show which direction that the breeding program should not take.
- This project provides farmers and agricultural consultants with access to greater genomic power. Some applications from this project may include better quality assurance, knowing when to plant a crop in order to change its environment, watching the soil temperature for when to plant a specific crop, or knowing how to change the soil microbiota in order to create a crop phenotype that is more resilient to stress.
Collaborations
Project 3.3 works closely with many other P2IRC Projects including:
- Project 1.2 Crop Phenometrics Platform and Project 1.3 Phenotyping the Plant Microbiome, which provide information to Project 3.3 on plant genomes and the soil-microbe complement, respectively.
- Project 2.2 Field-Based High-Throughput Phenotyping Mobile Systems for Crop Monitoring utilizes a semi-autonomous robot to collect environmental data analyzed in Project 3.3. Project 3.3 can then use this data to associate with phenotype data.
- Project 3.1 P2IRC Cloud assists in making computations faster, data storage, and sharing Project 3.3’s computational workload.
- Project 3.2 Data Analysis for Rapid Plant Phenotyping provides environmental information, phenotypic information, and interpretation for rapid phenotyping.
- Project 3.4 Systems and Collaboration assists in the human-computer interaction and providing groupware, so that breeders understand the technologies produced and can interact with their data.
Research Team
Loading...
Researchers:
Longhai Li
Kevin Stanley
Nadeem Jamali
Ian McQuillian
Post-Doctoral Fellows:
Jinhong Shi
Yan Yan
Nisha Puthiyedth
Graduate Students:
Ellen Redlick
Xiaoying Wang
Xuguang Yang
Saman Rahbar
Shaun Kaufmann
Undergraduate Research Assistants:
Conor Lazarou
Katherine Jamieson
Umar Tung
Alexa Armitage
Michelle Brabant
Connor Burbridge
Lujie Duan


